

Strategic advances in HIV research
Photo illustration by Jeffrey C. Chase | Photos by Kathy F. Atkinson and courtesy of Juan Perilla and Diamond Light Source November 19, 2021
UD’s Juan Perilla and team produce high-precision model of HIV dynamics in collaboration with Oxford
University of Delaware researcher Juan Perilla is a computational scientist with the heart of a storyteller. But he doesn’t want to tell computer stories. He wants to tell stories about the life cycle of viruses.
Perilla has been studying the inner workings of viruses for years, including the structure of the human immunodeficiency virus (HIV) and how it exploits a protein found in human cells to sneak past immune system defenses.
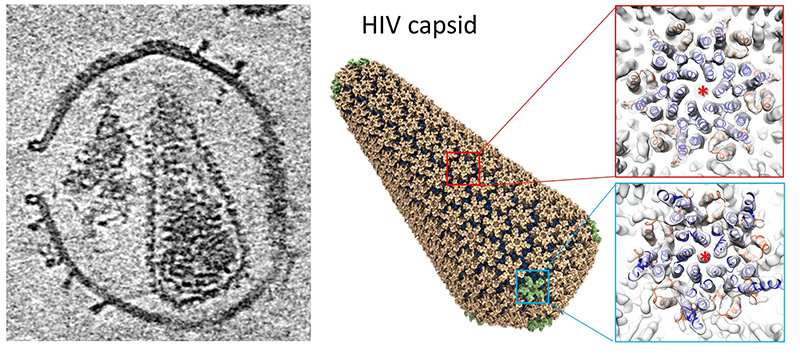
Now he has a new chapter to add. In a collaboration with Peijun Zhang, professor of structural biology at the University of Oxford, Perilla and two of his students have developed a computer model of the HIV shell — known as a capsid — that shows the whole 77-million atom system, how it moves and interacts with other cells.
Understanding this structure and especially its interactions with other cells could lead to new medicines to fight the virus that causes AIDS (acquired immunodeficiency syndrome). That would be good news for the estimated 38 million people living with HIV around the world.
A paper describing this work was published Friday, Nov. 19, in the journal Science Advances.
Perilla, assistant professor of chemistry and biochemistry at UD, and Zhang have known each other for at least 10 years, he said. This collaboration drew on the expertise of both of their labs.
Zhang and her team at Oxford used a new approach and state-of-the-art technology to show that perforating the membrane of an HIV particle allowed proteins and small molecules to reach its capsid, where the virus’s genome is stored. They then investigated how the capsid interacted with two specific molecules — one called CypA (cyclophilin A), the other IP6 (inositol hexakisphosphate). Those interactions are essential to the spread of the virus.
Perilla and his team used the data from the Oxford lab and supercomputers to build their model.
“This has atomistic precision,” Perilla said. “The ions, the cations, all of the hydrogens — it’s an atomistic view of the process, not an approximation. It’s as accurate as it gets. And we are able to tell the story from a biological point of view.”

“Free-energy” calculations are key to the model. These take all kinds of molecular dynamics into consideration, including potential of mean force (PMF), which is something like buoyancy or magnetism, sometimes drawing particles in, sometimes pushing them away. These dynamics are critical to the value of any model.
Some calculations were developed by Tanya Nesterova, an undergraduate student who had a double major in chemistry and applied mathematics. She has since graduated from UD and is now in a doctoral program at Johns Hopkins University. Also key to this work was Chaoyi Xu, a doctoral student in Perilla’s lab who has successfully defended his dissertation and now works as a computational chemist in the pharmaceutical industry.
“The combination of theory and experiments gives us an opportunity to train these very talented students,” Perilla said. “And this requires a very special kind of student — someone who studies physics, computing and math and wants to do something as far-fetched as biology. It’s hard to cross that barrier. I’ve been extremely privileged to work with these people.”
The model shows important targets that drug developers could explore, Perilla said.
“This is essential,” he said. “Millions of years of evolution have driven the virus in this direction.”
And this model could provide an important path forward for addressing other viruses, Zhang said.
“In collaboration with Prof. Juan Perilla’s group at the University of Delaware, using information derived from electron tomography, we also built an atomistic model of the whole HIV capsid which could serve as a blueprint for the development of capsid-targeting antivirals,” she said. “The perforation on the enveloped virus membrane also provides a novel approach to study host-virus interactions for other viral systems.”
Perilla and his lab have extensive experience in virus studies, including HIV, COVID-19, hepatitis B, and Ebola.
“I’m looking forward to seeing how my students develop,” Perilla said. “They’re the future. They’ve been through a pandemic and been trained through HIV. They are the ones who are going to tackle the problems when I’m old and I have no doubt they’ll succeed.”
Many questions remain about what these molecules and metabolites are doing, but this work adds to the narrative.
“Even though HIV is one of the most studied viruses, there are so many questions that remain unclear or under debate about how HIV infects and replicates in the cell,” said Xu. “By investigating HIV in detail, it not only gives us new knowledge about fighting the ongoing HIV/AIDS epidemic, but also insights about the replication cycles of other viruses.”
The collaboration was an amazing experience, Xu said.
“We are very happy to see that our collaboration made some impacts that we would not achieve individually,” he said. “I personally also learned a lot by working with the researchers and students from other labs and different backgrounds.”
Perilla hopes many more students will be drawn to this work.
“The U.S. has extremely strong, well-trained people who can produce pharmaceuticals,” Perilla said. “But a very thin part of the population devotes their lives to this work. We need more opportunities to bring people to science and have them be successful in science. The talent is out there. We just need more people.”
In addition to Zhang, this collaborative work included scientists from the Electron Bio-Imaging Centre (eBIC), which she directs, and partners at the University of Pittsburgh School of Medicine, the School of Medical Sciences in Sydney, Australia, the University of Melbourne and St. Vincent’s Institute of Medical Research in Victoria, Australia. The lead authors of the Science Advances article were Tao Ni and Yanan Zhu of the University of Oxford.
Support for the work came from multiple sources, including the National Institutes of Health, the Wellcome Trust, the United Kingdom’s Biotechnology and Biological Sciences Research Council and the Australian Research Council.
About the Researcher
Juan Perilla is an assistant professor in the Department of Chemistry and Biochemistry at the University of Delaware. He earned his doctorate at Johns Hopkins University and did postdoctoral work at the University of Illinois at Urbana-Champaign before joining the UD faculty.
Contact Us
Have a UDaily story idea?
Contact us at ocm@udel.edu
Members of the press
Contact us at 302-831-NEWS or visit the Media Relations website

